UMAP
The UMAP dimensionality reduction tool offers a workflow for dimensionality reduction and clustering that is particularly useful for high dimensional data, such as hyperspectral images. The tool uses UMAP (Uniform Manifold Approximation and Projection) to reduce the dimensionality of the data, followed by HDBSCAN (Heirarchical Density Based Spatial Clustering of Applications with Noise).
Warning
UMAP requires access to Compute.
Tip
Mathematical details for both UMAP and HDBSCAN can be found in the original papers: UMAP and HDBSCAN.
Selecting data
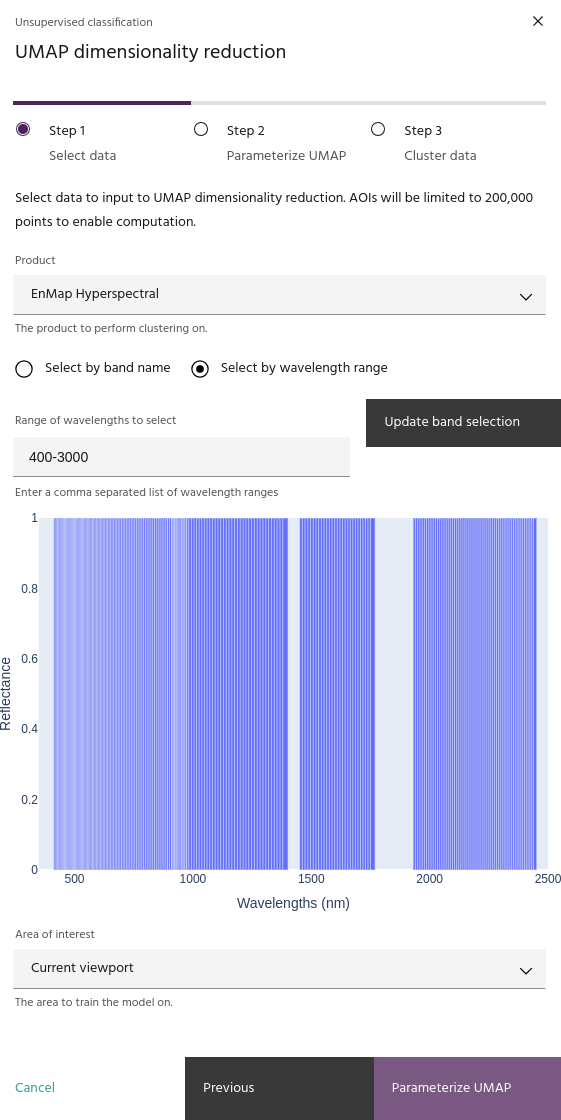
The UMAP dialog is broken into three steps. The first step allows you to
select the product, bands
and AOI for analysis. When you have selected
the data you want to analyze, click Parameterize UMAP to move to the
next step.
Warning
Selected data cannot have any fully masked bands. If necessary, use the data statistical analysis tool to clean data before running UMAP.
Note
Data will be limited to 50,000 points to manage runtimes.
Parameterize UMAP
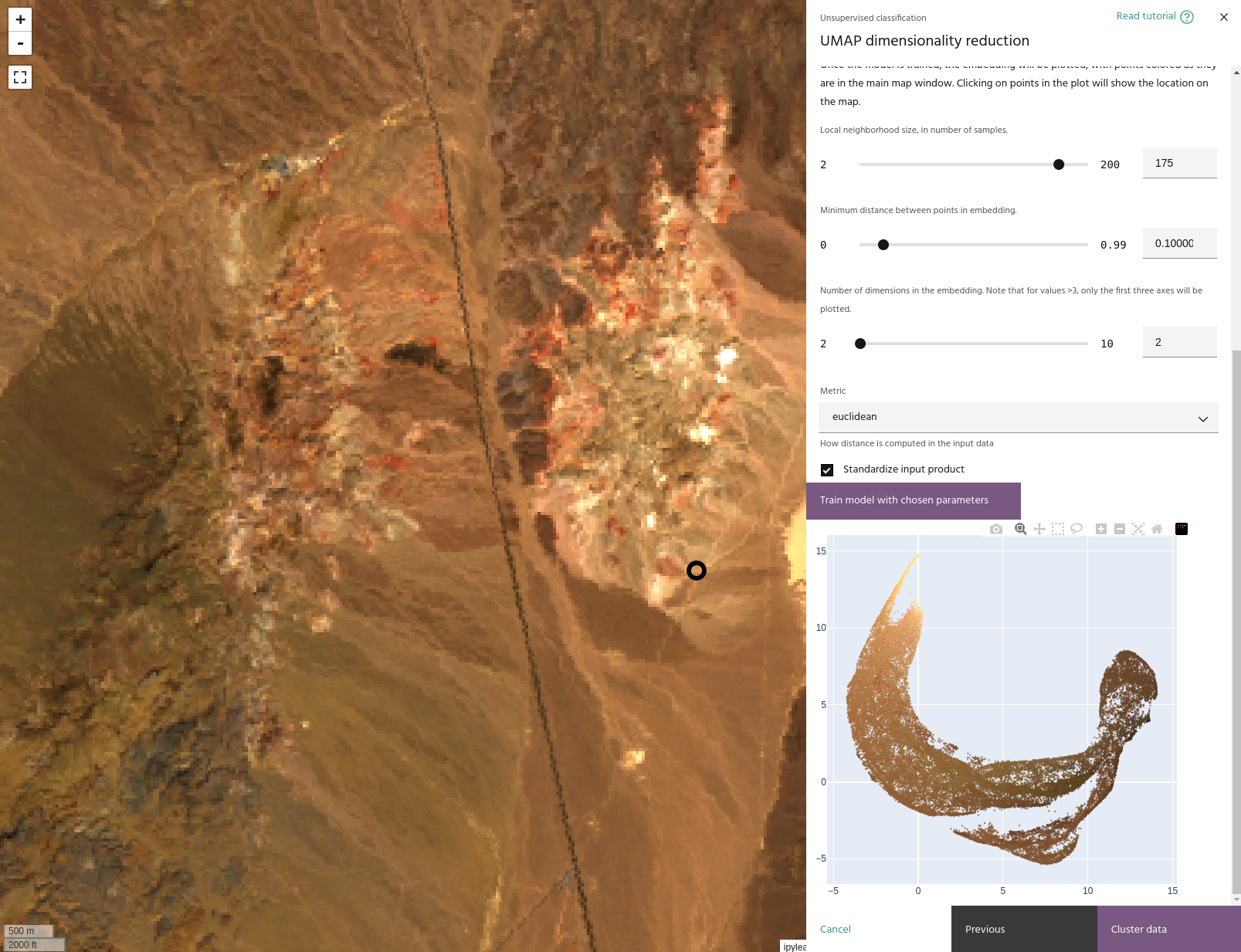
UMAP works by computing a manifold of the input data and projecting that manifold
to a lower dimensional space. Parameterizing UMAP, therefore, involves determining
how that manifold is built (the neighborhood size and
distance metric used), how many dimensions to
use in the projection, and how clustered the low dimensional
projection is. The second step of the dialog allows manipulation of these parameters
and provides a plot of the embedding for analysis. When you have set parameters to your satisfaction, click Train model with chosen parameters to train the model and generate
a plot of the embedding, and when you are satisfied with your model click Cluster data
to move to the clustering step.
Tip
More details on parameters can be found here
Tip
Points in the embedding plot will be colored according to the colors of the layer on the map. Clicking on a point in the plot will show its location on the map.
Note
The first time training a model with a given choice of metric, the K nearest neighbors will be computed and stored. This enables faster parameter testing, without needing to run this step each time.
Local neighborhood size
The number of points that UMAP will use when building the data manifold. This parameter allows a balance between local and global structure of the data in the embedding - low values will focus on local structure, and large values will focus on global structure.
Minimum distance
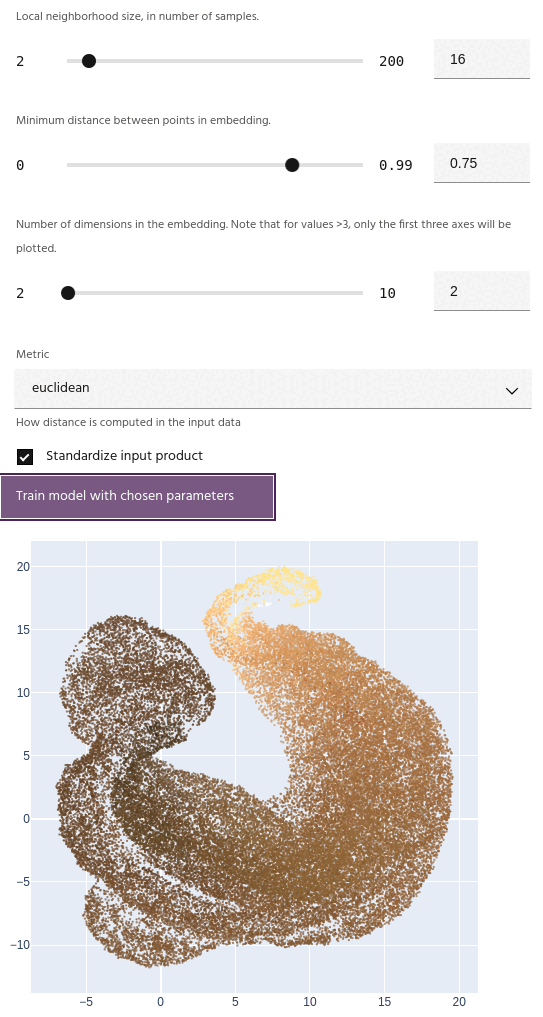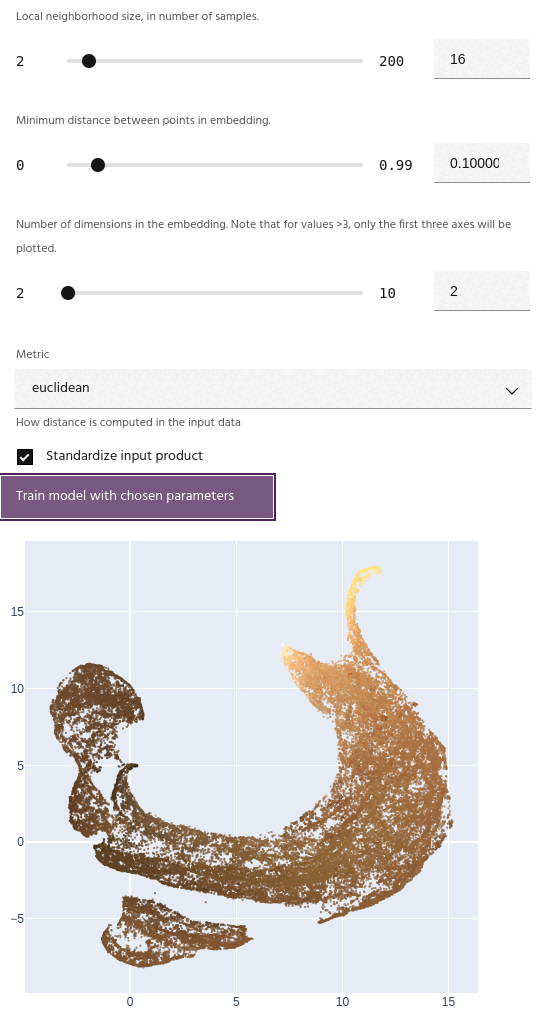
The minimum distance between points in the final embedding. Lower values will create a clumpier embedding, while larger values will spread points farther apart.
Tip
This parameter has a strong effect on the number of clusters generated - small values tend to create fewer large clusters, and large values create more small clusters.
Number of dimensions
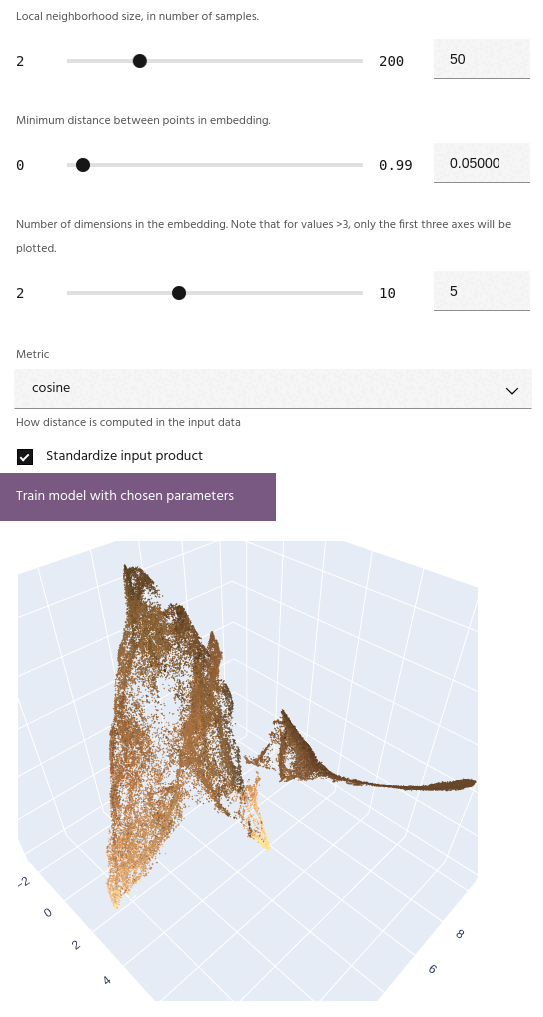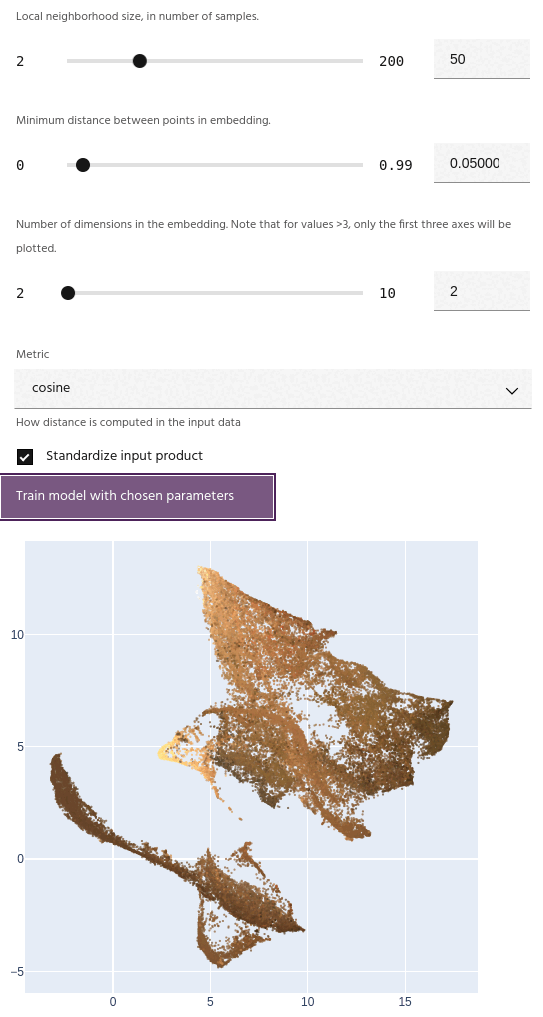
Number of dimensions for the embedding. Unlike simpler embedding algorithms such as T-SNE, UMAP can successfully embed into more than three dimensions.
Note
If the number of dimensions is greater than 3, the embedding plot will show the first three axes.
Metric
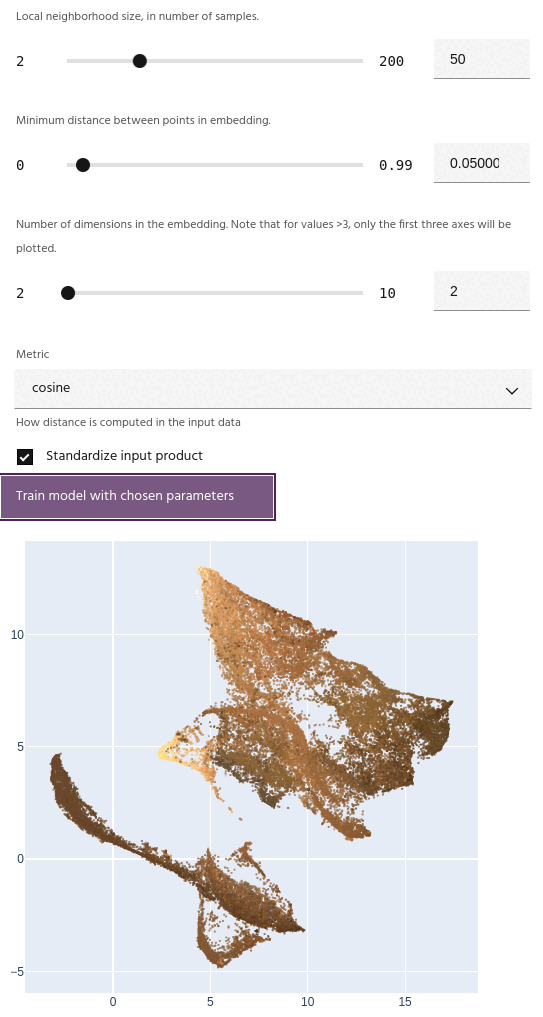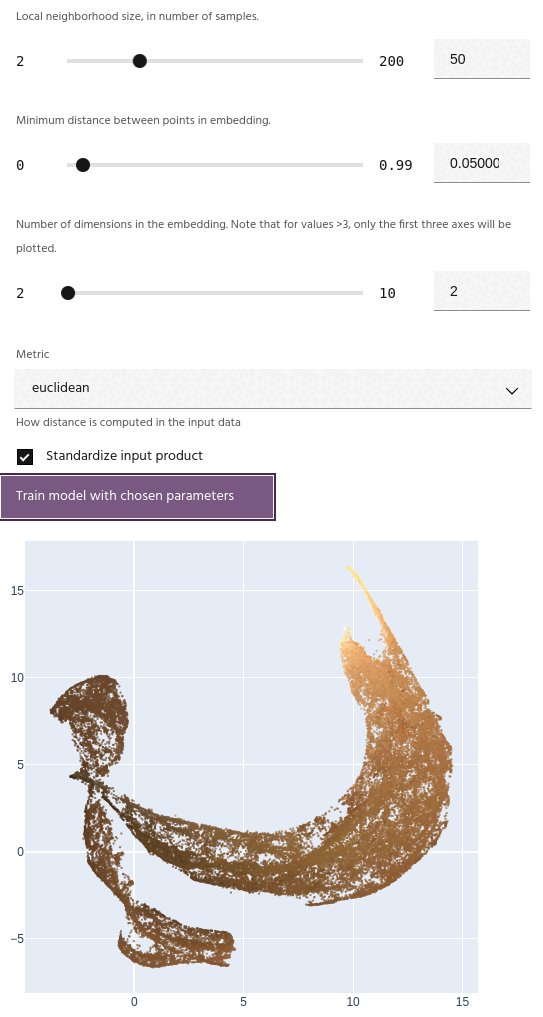
The metric for computing distances in the input data. Available metrics are:
- Euclidean: Simple Euclidean distance between points.
- Manhattan: Manhattan, or "Taxi cab" distance, measures the sum of the distance along each axis.
- Chebyshev: The Chebyshev metric defines the distance between points as the greatest of their differences along any axis.
- Cosine: Cosine distance is the cosine of the angle between the vectors.
- Correlation: Correlation distance between the vectors, measuring the dependence between them.
Tip
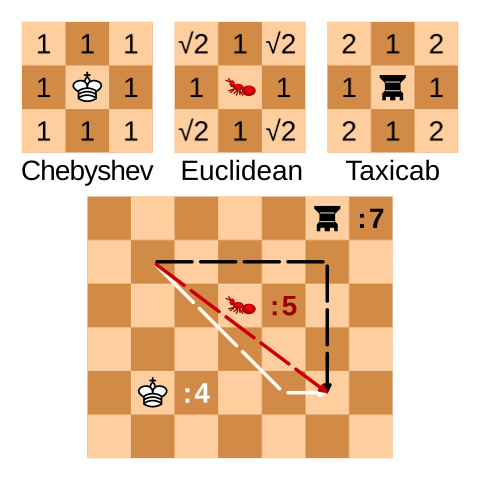
Euclidean, Manhattan, and Chebyshev distances can easily be visualized by their behavior on a chess board. Chebyshev distance corresponds to the number of moves a King makes getting from one square to the other, Manhattan distance behaves like a Rook, and Euclidean distance is an ant moving without regard for the board:
Embeddings plot
After training a model, the embedding will be plotted on either a 2D or 3D scatterplot,
depending on the choice of dimension. Points on the plot
will be colored the same as they are on the map, and clicking on plotted point will
show the location on the map in a vector layer called
selected points.
Note
The axes for the embedding are dimensionless, and simply represent a new spatial position for the input points relative to all of the others.
Cluster data
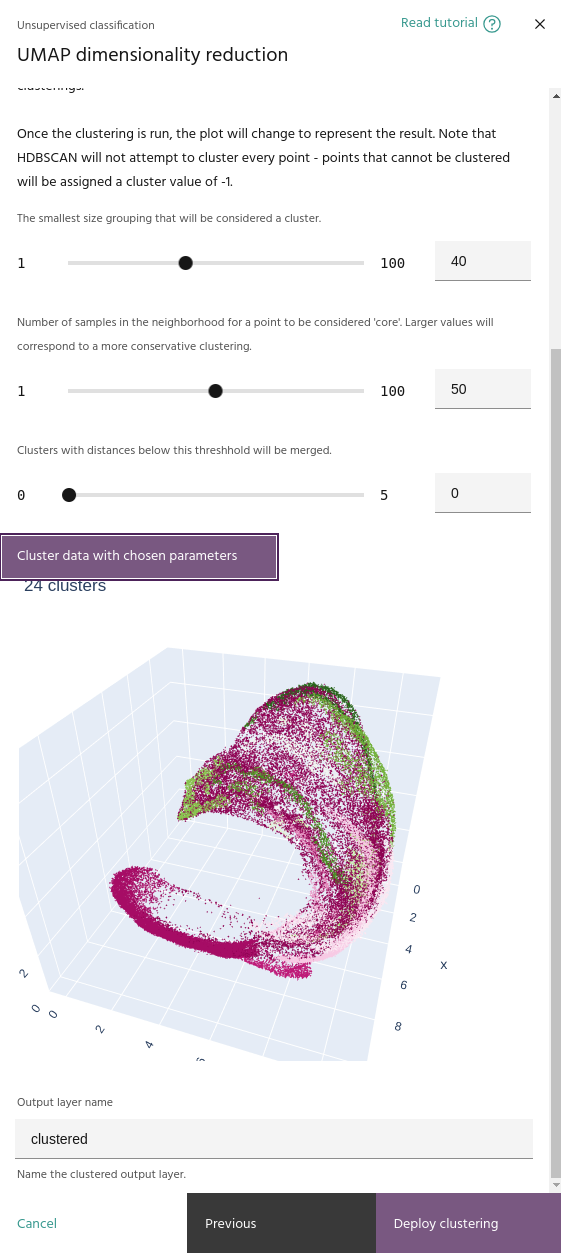
Once the low dimensional manifold is built, the UMAP tool uses HDBSCAN to cluster
the data in an unsupervised manner. HDBSCAN is a better clustering algorithm than
k-means for the unique shapes that are generated with UMAP, as it
can find clusters with varied shapes, whereas k-means builds clusters as balls.
Use the Cluster data with chosen parameters button to run the clustering on your
embedded data.
Tip
HDBSCAN will not attempt to cluster outlier points, and will instead classify them separately as noise.
Tip
When you run a clustering, the embeddings plot will change to show the clusters instead of the pixel RGBs.
Minimum cluster size
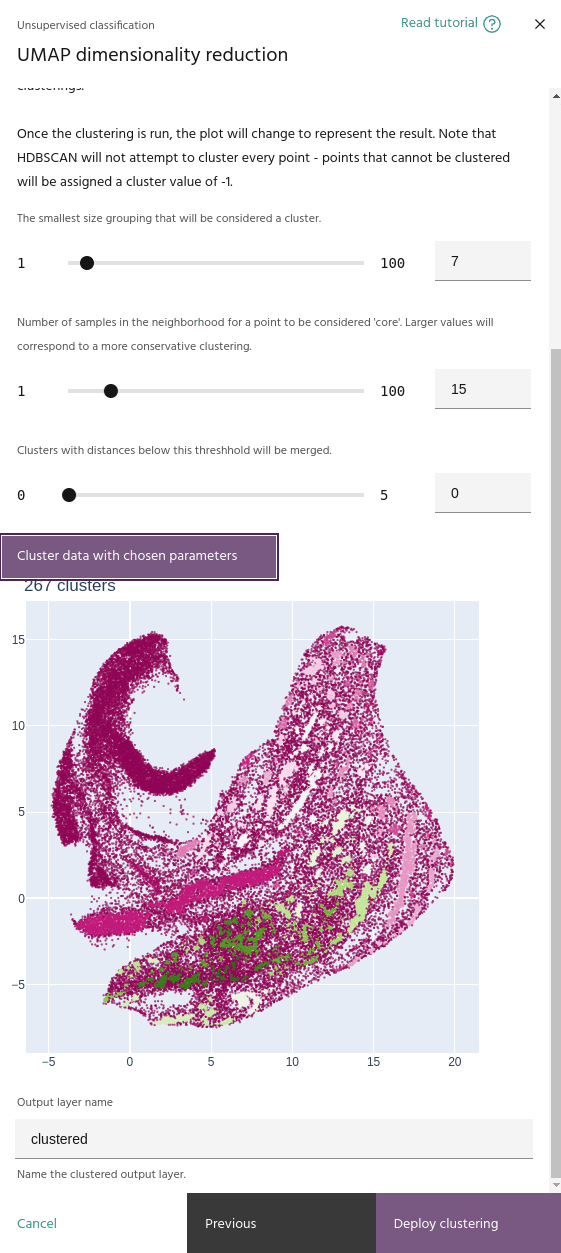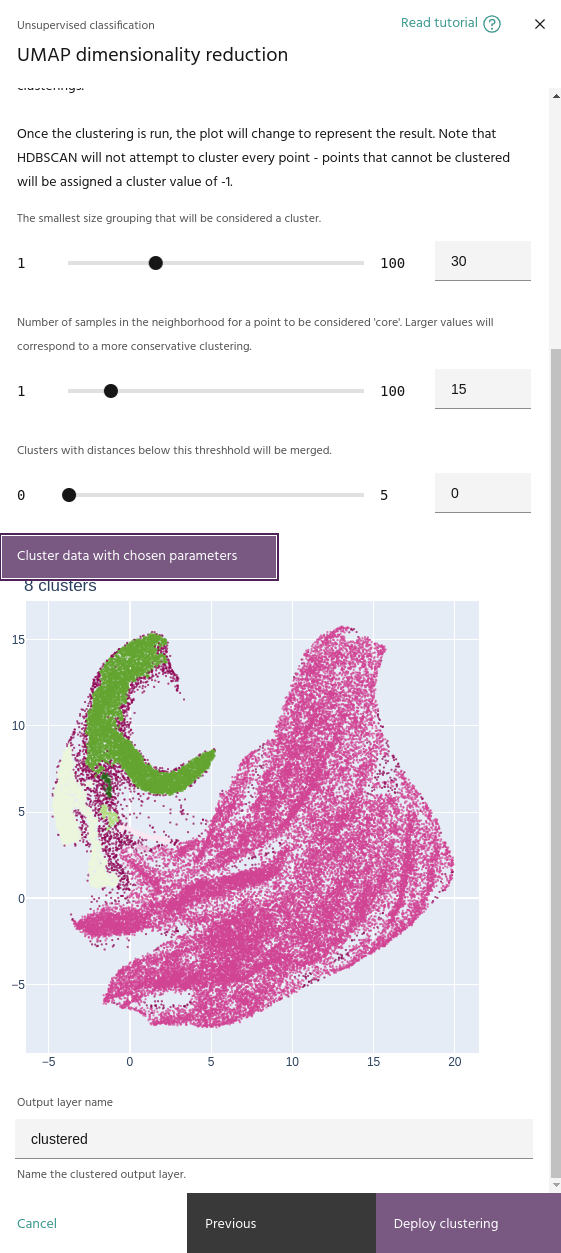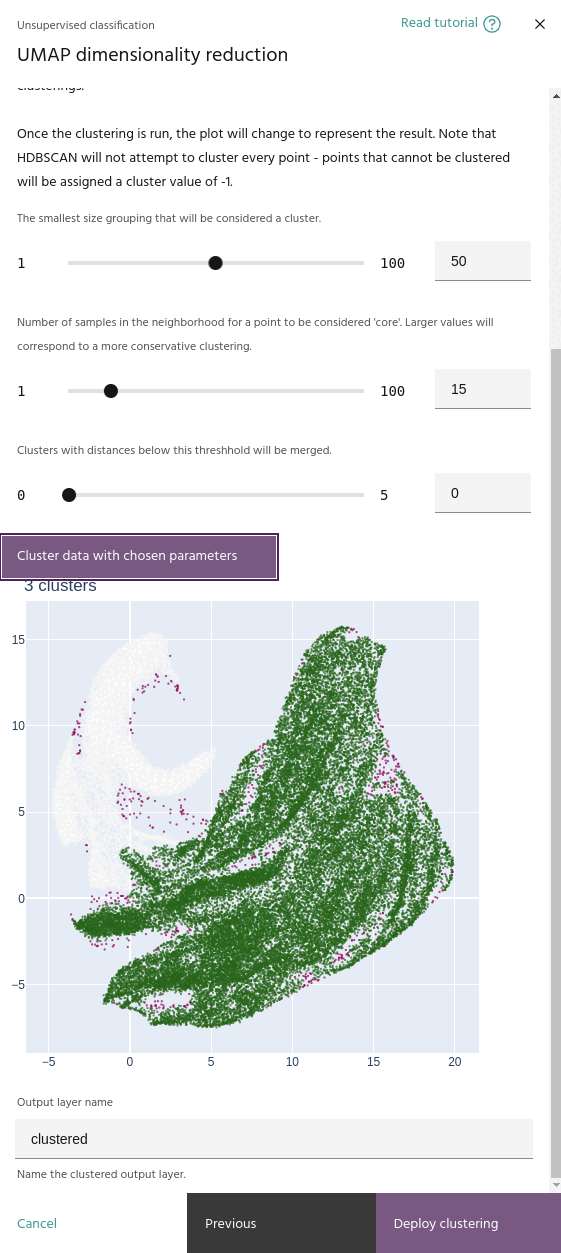
The smallest number of points that can be considered a cluster. Lower values tend to create more clusters.
Minimum number of samples
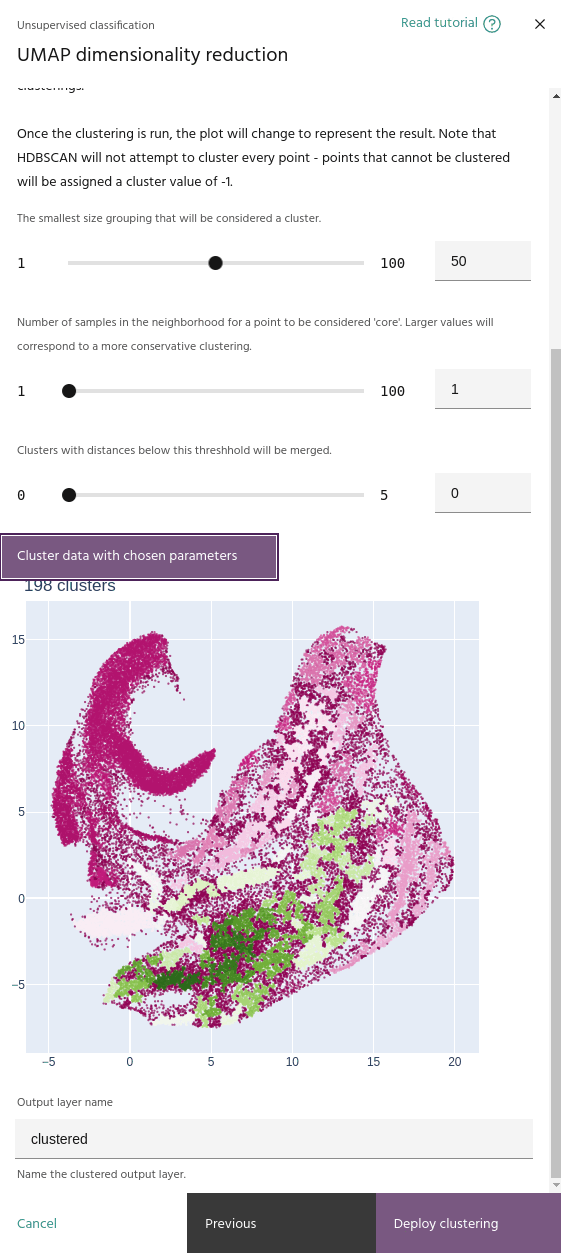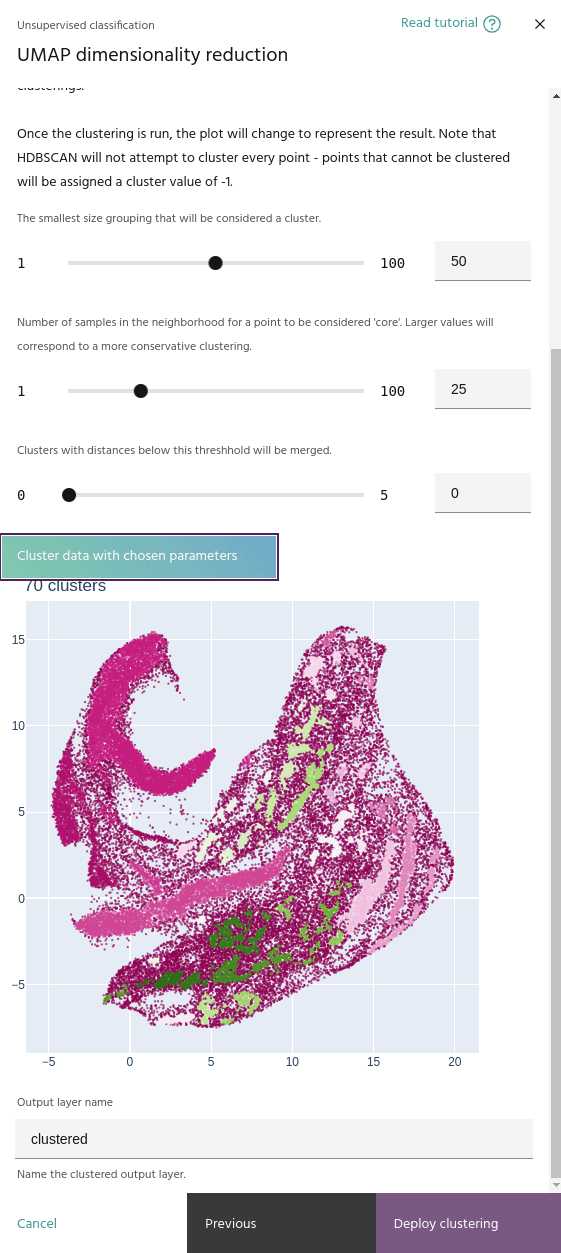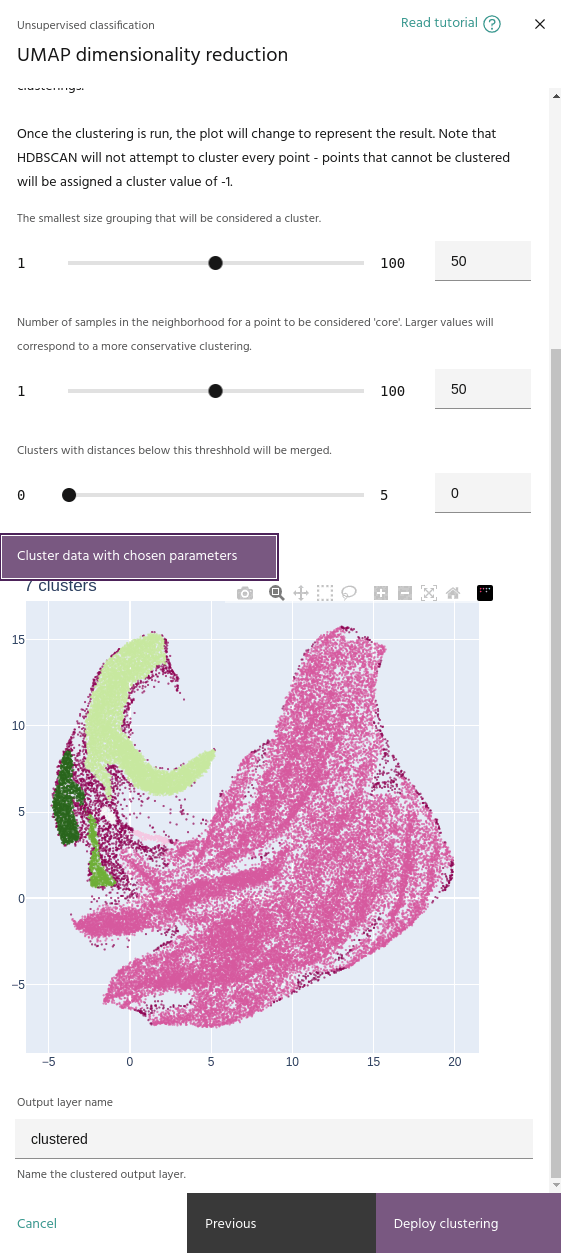
The minimum number of samples in the neighborhood of a point for it to be considered a "core" sample, and therefore considered for clustering instead of noise.
Cluster selection epsilon
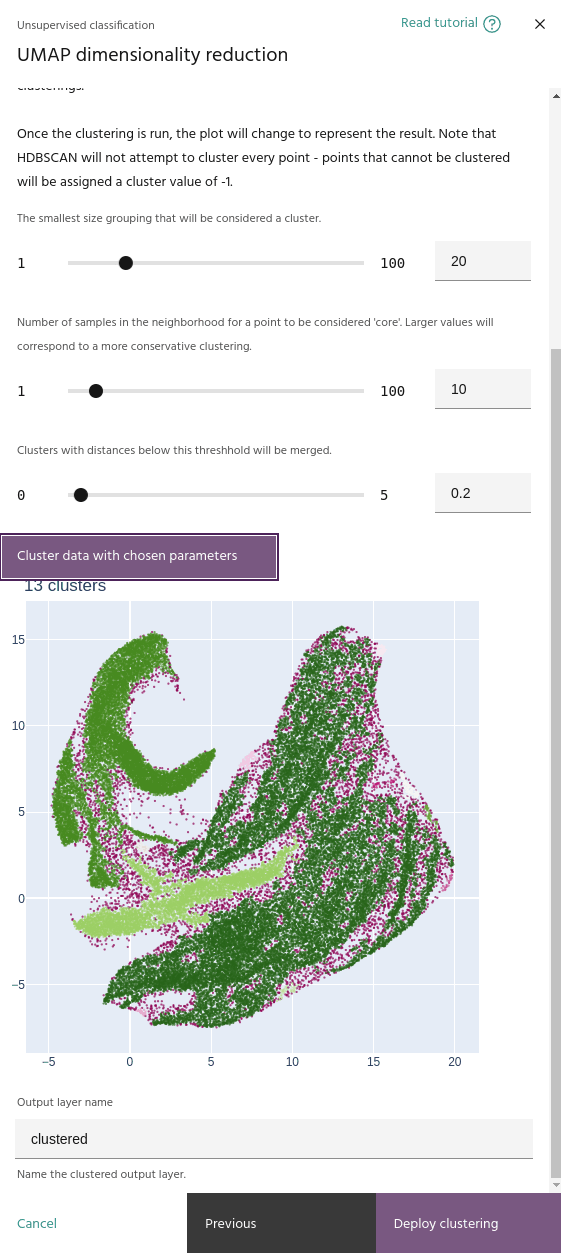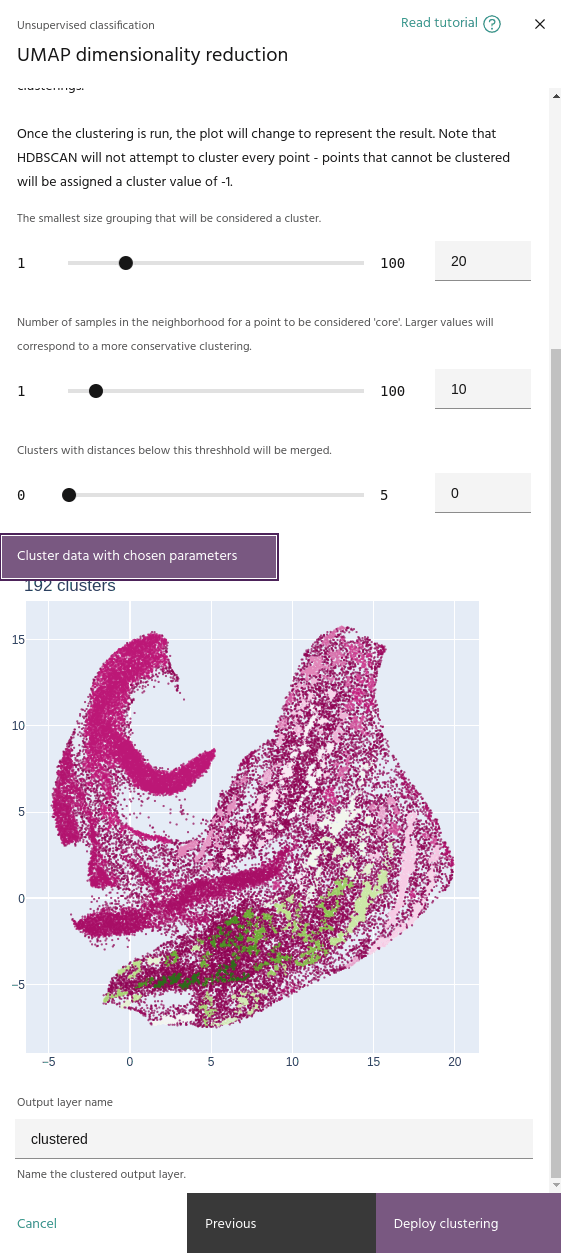
This parameter merges clusters that are closer than this value. This will help if your clustering contains a large number of small, nearby clusters.
Deploy clustering
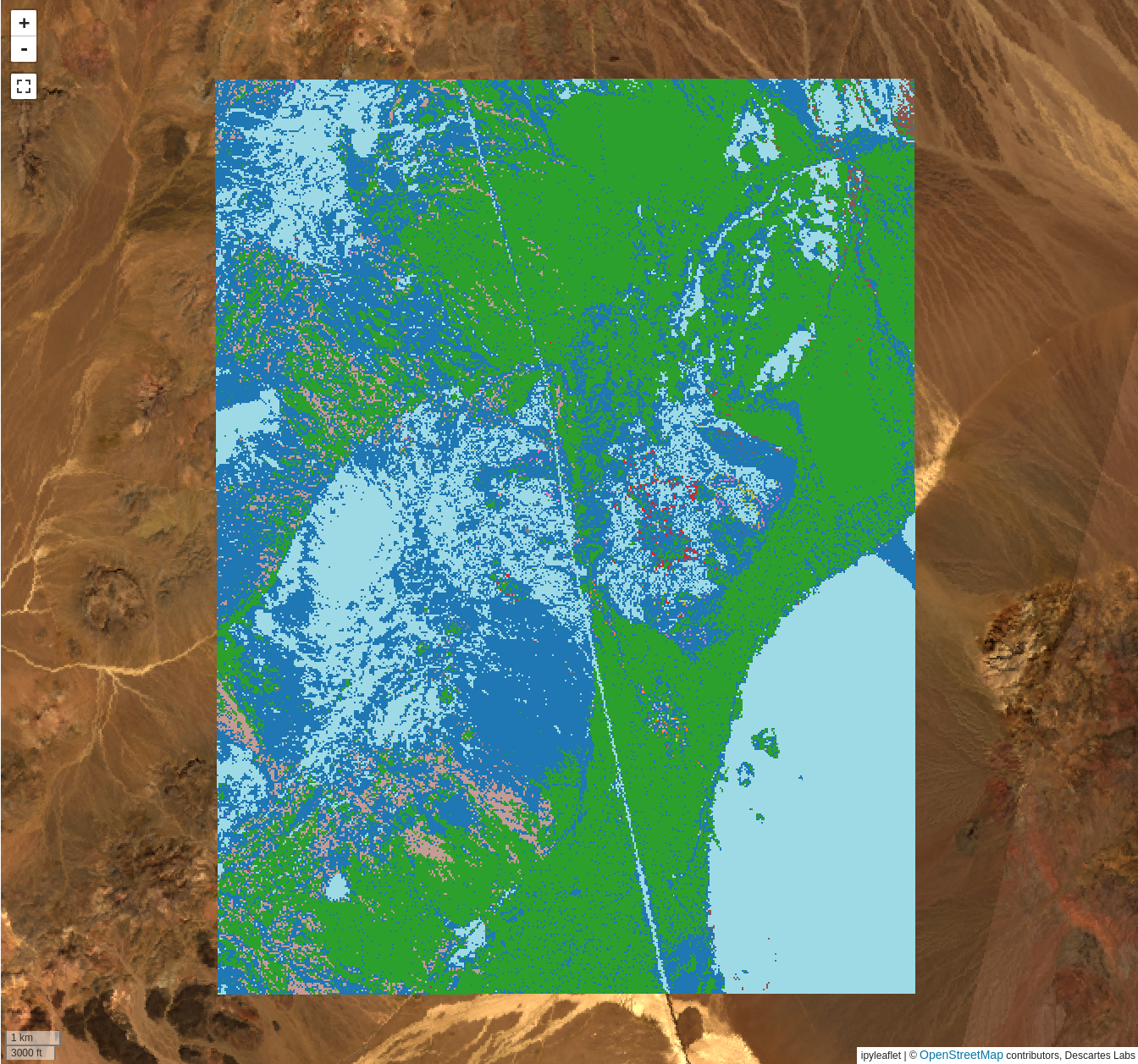
When you are happy with the clustering parameters, click the Deploy clustering.
This will deploy a Compute
function that will apply the UMAP and HDBSCAN models over your chosen AOI, and
add the result to the map.
Tip
Progress on the Compute task can be found here.
Note
Images for the clustered result will appear on the map as they are completed, but you may need to override the map's cache to see the latest images. This can be accomplished by moving around the map, or changing the settings of the layer.
Warning
Only the AOI where data were selected will be clustered. As the models are build using these data, applying the models to areas outside this AOI is not valid.